i | Adj_anova(i) |
0 | Degrees of freedom for groups after covariates. |
1 | Degrees of freedom for covariates after groups. |
2 | Sum of squares for groups after covariates. |
3 | Sum of squares for model (groups and covariates combined). |
4 | F -statistic for groups. |
5 | F -statistic for covariates. |
6 | p-value for groups. |
7 | p-value for covariates. |
i | testpl(i) |
0 | Extra degrees of freedom for model not assuming parallelism. |
1 | Degrees of freedom for error for model not assuming parallelism. |
2 | Degrees of freedom for error for model assuming parallelism. |
3 | Extra sum of squares for model not assuming parallelism. |
4 | Sum of squares for error for model not assuming parallelism. |
5 | Sum of squares for error for model assuming parallelism. |
6 | Mean square for testpl(0). |
7 | Mean square for testpl(1). |
8 | F-statistic. |
9 | p-value. |
Column | Description |
0 | Number of non-missing cases |
1 through ncov | Covariate means. |
ncov + 1 | Response mean. |
ncov + 2 | Response mean adjusted assuming parallelism. |
j | Anova_tables(i,j) |
0 | Degrees of freedom for regression model (covariates). |
1 | Degrees of freedom for error. |
2 | Total (corrected) degrees of freedom. |
3 | Sum of squares for regression model. |
4 | Sum of squares for error. |
5 | Total (corrected) sum of squares. |
6 | Model mean square. |
7 | Error mean square. |
8 | F-statistic. |
9 | p-value. |
10 | R2 (in percent). |
11 | Adjusted R2 (in percent). |
12 | Error standard deviation. |
13 | Overall response mean. |
14 | Coefficient of variation (in percent). |
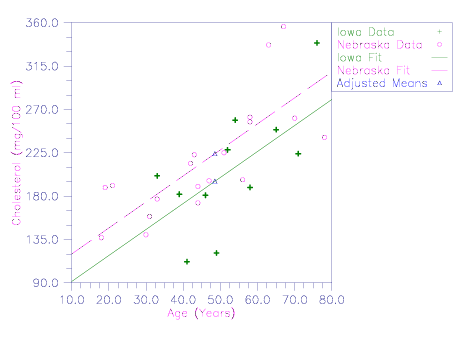
Iowa | Nebraska | |||
Age | Cholesterol | Age | Cholesterol | |
46 | 181 | 18 | 137 | |
52 | 228 | 44 | 173 | |
39 | 182 | 33 | 177 | |
65 | 249 | 78 | 241 | |
54 | 259 | 51 | 225 | |
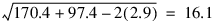
PRO t_ancovar_ex1
ncov=1
ngroup=2
ni = [11, 19]
nobs = TOTAL(ni)
y = $
[181.0, 228.0, 182.0, 249.0, 259.0,$
201.0, 121.0, 339.0, 224.0, 112.0,$
189.0, 137.0, 173.0, 177.0, 241.0,$
225.0, 223.0, 190.0, 257.0, 337.0,$
189.0, 214.0, 140.0, 196.0, 262.0,$
261.0, 356.0, 159.0, 191.0, 197.0]
x = $ ; Should be nobs x ncov.
[46.0, 52.0, 39.0, 65.0, 54.0,$
33.0, 49.0, 76.0, 71.0, 41.0,$
58.0, 18.0, 44.0, 33.0, 78.0,$
51.0, 43.0, 44.0, 58.0, 63.0,$
19.0, 42.0, 30.0, 47.0, 58.0,$
70.0, 67.0, 31.0, 21.0, 56.0]
aov = ANCOVAR(ni, y, x,$
Testpl=testpl, $
Xymean=xymean, $
Covmeans=covm)
PRINT," * * * ANALYSIS OF VARIANCE * * * "
PRINT," Sum of Mean Prob of"
PRINT,"Source DF Squares Square Overall F"+ $
" Larger F"
PRINT,"Model ",STRING(aov(0),Format="(f3.0)")," ",$
STRING(aov(3),Format="(f10.2)")," ",$
STRING(aov(6),Format="(f9.2)")," ",$
STRING(aov(8),Format="(f8.2)")," ",$
STRING(aov(9),Format="(f8.6)")
PRINT,"Error ",STRING(aov(1),Format="(f3.0)")," ",$
STRING(aov(4),Format="(f10.2)")," ",$
STRING(aov(7),Format="(f9.2)")
PRINT,"Total ",STRING(aov(2),Format="(f3.0)")," ",$
STRING(aov(5),Format="(f10.2)")
PRINT,""
PRINT," * * * TEST FOR PARALLELISM * * * "
PRINT," Sum of Mean F Prob of"
PRINT,"SOURCE DF Squares Square TEST Larger F"
PRINT,"Extra due to"
PRINT,"Nonparallelism ",STRING(testpl(0), $
Format="(f3.0)"),"",$
STRING(testpl(3),Format="(f10.2)")," ",$
STRING(testpl(6),Format="(f7.2)")," ",$
STRING(testpl(8),Format="(f7.5)")," ",$
STRING(testpl(9),Format="(f8.4)")
PRINT,"Extra Assuming"
PRINT,"Nonparallelism ",STRING(testpl(1),$
Format="(f3.0)"),"",$
STRING(testpl(4),Format="(f10.2)")," ",$
STRING(testpl(7),Format="(f7.2)")
PRINT,"Error Assuming"
PRINT,"Parallelism ",STRING(testpl(2),$
Format="(f3.0)"),"",$
STRING(testpl(5),Format="(f10.2)")
PRINT,""
PRINT," XY Mean Matrix"
PRINT," 1 2 3 4"
FOR i=0L, ngroup DO BEGIN
PRINT,STRTRIM(i+1,2)," ",$
STRING(xymean(i,0),Format="(f6.1)")," ",$
STRING(xymean(i,1),Format="(f6.1)")," ",$
STRING(xymean(i,2),Format="(f6.1)")," ",$
STRING(xymean(i,3),Format="(f6.1)")
ENDFOR
PRINT,""
PRINT," Var./Covar. Matrix of Adjusted Group Means"
PRINT," 1 2"
FOR i=0L, ngroup-1 DO BEGIN
PRINT," ",STRTRIM(i+1,2)," ",$
STRING(covm(i,0),Format="(f6.1)")," ",$
STRING(covm(i,1),Format="(f6.1)")
ENDFOR
END
* * * ANALYSIS OF VARIANCE * * *
Sum of Mean Prob of
Source DF Squares Square Overall F Larger F
Model 2. 54432.76 27216.38 14.97 0.000042
Error 27. 49103.90 1818.66
Total 29. 103536.66
* * * TEST FOR PARALLELISM * * *
Sum of Mean F Prob of
SOURCE DF Squares Square TEST Larger F
Extra due to
Nonparallelism 1. 709.04 709.04 0.38093 0.5425
Extra Assuming
Nonparallelism 26. 48394.87 1861.34
Error Assuming
Parallelism 27. 49103.90
XY Mean Matrix
1 2 3 4
1 11.0 53.1 207.7 195.5
2 19.0 45.9 217.1 224.2
3 30.0 48.6 213.7 213.7
Var./Covar. Matrix of Adjusted Group Means
1 2
1 170.4 -2.9
2 -2.9 97.4
 |
Treatment 1 | Treatment 2 | Treatment 3 | Treatment 4 | ||||||||
Age | Wt. | Gain | Age | Wt. | Gain | Age | Wt. | Gain | Age | Wt. | Gain |
78 | 61 | 1.40 | 78 | 74 | 1.61 | 78 | 80 | 1.67 | 77 | 62 | 1.40 |
90 | 59 | 1.79 | 99 | 75 | 1.31 | 83 | 61 | 1.41 | 71 | 55 | 1.47 |
94 | 76 | 1.72 | 80 | 64 | 1.12 | 79 | 62 | 1.73 | 78 | 62 | 1.37 |
71 | 50 | 1.47 | 75 | 48 | 1.35 | 70 | 47 | 1.23 | 70 | 43 | 1.15 |
99 | 61 | 1.26 | 94 | 62 | 1.29 | 85 | 59 | 1.49 | 95 | 57 | 1.22 |
PRO t_ancovar_ex2
ncov=2
ngroup=4
ni = [10, 10, 10, 10]
nobs = SUM(ni)
x1 = $
[78.0, 90.0, 94.0, 71.0, 99.0, 80.0, 83.0, 75.0, 62.0, 67.0,$
78.0, 99.0, 80.0, 75.0, 94.0, 91.0, 75.0, 63.0, 62.0, 67.0,$
78.0, 83.0, 79.0, 70.0, 85.0, 83.0, 71.0, 66.0, 67.0, 67.0,$
77.0, 71.0, 78.0, 70.0, 95.0, 96.0, 71.0, 63.0, 62.0, 67.0]
x2 = $
[61.0, 59.0, 76.0, 50.0, 61.0, 54.0, 57.0, 45.0, 41.0, 40.0,$
74.0, 75.0, 64.0, 48.0, 62.0, 42.0, 52.0, 43.0, 50.0, 40.0,$
80.0, 61.0, 62.0, 47.0, 59.0, 42.0, 47.0, 42.0, 40.0, 40.0,$
62.0, 55.0, 62.0, 43.0, 57.0, 51.0, 41.0, 40.0, 45.0, 39.0]
y = $
[1.40, 1.79, 1.72, 1.47, 1.26, 1.28, 1.34, 1.55, 1.57, 1.26,$
1.61, 1.31, 1.12, 1.35, 1.29, 1.24, 1.29, 1.43, 1.29, 1.26,$
1.67, 1.41, 1.73, 1.23, 1.49, 1.22, 1.39, 1.39, 1.56, 1.36,$
1.40, 1.47, 1.37, 1.15, 1.22, 1.48, 1.31, 1.27, 1.22, 1.36]
x=FLTARR(nobs,ncov)
; Set up covariate input matrix.
x(*,0) = x1
x(*,1) = x2
aov = ANCOVAR(ni, y, x, $
Testpl=testpl, $
Adj_anova=adj_aov, $
Xymean=xymean, $
Covmeans=covm)
PRINT,""
PRINT," * * * TEST FOR PARALLELISM * * * "
PRINT," Sum of Mean F Prob of"
PRINT,"SOURCE DF Squares Square TEST"+$
" Larger F"
PRINT,"Extra due to"
PRINT,"Nonparallelism ",STRING(testpl(0),$
Format="(f3.0)")," ",$
STRING(testpl(3),Format="(f10.2)")," ",$
STRING(testpl(6),Format="(f7.2)")," ",$
STRING(testpl(8),Format="(f7.5)")," ",$
STRING(testpl(9),Format="(f7.4)")
PRINT,"Extra Assuming"
PRINT,"Nonparallelism ",STRING(testpl(1),$
Format="(f3.0)"),"",$
STRING(testpl(4),Format="(f10.2)")," ",$
STRING(testpl(7),Format="(f7.2)")
PRINT,"Error Assuming"
PRINT,"Parallelism ",STRING(testpl(2),$
Format="(f3.0)"),"",$
STRING(testpl(5),Format="(f10.2)")
PRINT,""
PRINT," * * * ANALYSIS OF VARIANCE * * * "
PRINT," Sum of Mean Prob of"
PRINT,"Source DF Squares Square Overall F"+$
" Larger F"
PRINT,"Model ",STRING(aov(0),Format="(f3.0)")," ",$
STRING(aov(3),Format="(f10.5)")," ",$
STRING(aov(6),Format="(f9.5)")," ",$
STRING(aov(8),Format="(f9.5)")," ",$
STRING(aov(9),Format="(f8.6)")
PRINT,"Error ",STRING(aov(1),Format="(f3.0)")," ",$
STRING(aov(4),Format="(f10.5)")," ",$
STRING(aov(7),Format="(f9.5)")
PRINT,"Total ",STRING(aov(2),Format="(f3.0)")," ",$
STRING(aov(5),Format="(f10.5)")
PRINT,""
PRINT,""
PRINT," * * * ADJUSTED ANALYSIS OF VARIANCE * * * "
PRINT," Sum of F Prob of"
PRINT,"Source DF Squares TEST Larger F"
PRINT,"Groups after Covariates ",STRING(adj_aov(0),$
Format="(f3.0)"),"",$
STRING(adj_aov(2),Format="(f10.2)")," ",$
STRING(adj_aov(4),Format="(f5.2)")," ",$
STRING(adj_aov(6),Format="(f7.5)")
PRINT,"Covariates after Groups ",STRING(adj_aov(1),$
Format="(f3.0)"),"",$
STRING(adj_aov(3),Format="(f10.2)")," ",$
STRING(adj_aov(5),Format="(f5.2)")," ",$
STRING(adj_aov(7),Format="(f7.5)")
PRINT,""
PRINT," * * * GROUP MEANS * * * "
PRINT,"GROUP | Unadjusted | Adjusted | Std. Error"
FOR i=0L, ngroup-1 DO BEGIN
stderr = SQRT(covm(i,i))
PRINT, STRTRIM(i+1,2)," | ", $
STRING(xymean(i, ngroup-1),Format="(f7.4)")," | ",$
STRING(xymean(i, ngroup ),Format="(f7.4)")," | ",$
STRING(stderr,Format="(f7.4)")
ENDFOR
PRINT,""
PRINT," * * * STUDENT-T MULTIPLE COMPARISONS * * * "
PRINT," GROUPS | DIFF | Std. Error | Student-t | P-Value"
FOR i=0L, ngroup-1 DO BEGIN
FOR j=i+1, ngroup-1 DO BEGIN
delta = xymean(i,ngroup) - $
xymean(j,ngroup)
stderr = SQRT(covm(i,i)+covm(j,j)- $
2.0*covm(i,j))
t = delta/stderr;
df = xymean(i,0)+xymean(j,0)-2
pvalue = 1.0 - TCDF(t, df)
PRINT, STRTRIM(i+1,2)," vs ",STRTRIM(j+1,2)," | ",$
STRING(delta,Format="(f7.4)")," | ",$
STRING(stderr,Format="(f7.4)")," | ",$
STRING(t,Format="(f7.3)")," | ",$
STRING(pvalue,Format="(f7.5)")
ENDFOR
ENDFOR
END
* * * TEST FOR PARALLELISM * * *
Sum of Mean F Prob of
SOURCE DF Squares Square TEST Larger F
Extra due to
Nonparallelism 6. 0.05 0.01 0.35534 0.9007
Extra Assuming
Nonparallelism 28. 0.62 0.02
Error Assuming
Parallelism 34. 0.67
* * * ANALYSIS OF VARIANCE * * *
Sum of Mean Prob of
Source DF Squares Square Overall F Larger F
Model 5. 0.35252 0.07050 3.57640 0.010491
Error 34. 0.67026 0.01971
Total 39. 1.02278
* * * ADJUSTED ANALYSIS OF VARIANCE * * *
Sum of F Prob of
Source DF Squares TEST Larger F
Groups after Covariates 3. 0.17 2.90 0.04931
Covariates after Groups 2. 0.17 4.44 0.01939
* * * GROUP MEANS * * *
GROUP | Unadjusted | Adjusted | Std. Error
1 | 1.4640 | 1.4614 | 0.0448
2 | 1.3190 | 1.3068 | 0.0446
3 | 1.4450 | 1.4429 | 0.0447
4 | 1.3250 | 1.3418 | 0.0449
* * * STUDENT-T MULTIPLE COMPARISONS * * *
GROUPS | DIFF | Std. Error | Student-t | P-Value
1 vs 2 | 0.1546 | 0.0630 | 2.455 | 0.01225
1 vs 3 | 0.0185 | 0.0637 | 0.290 | 0.38750
1 vs 4 | 0.1196 | 0.0638 | 1.875 | 0.03854
2 vs 3 | -0.1362 | 0.0632 | -2.153 | 0.97743
2 vs 4 | -0.0350 | 0.0638 | -0.549 | 0.70528
3 vs 4 | 0.1011 | 0.0631 | 1.602 | 0.06330