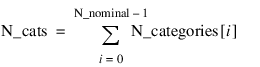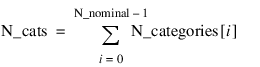GA_RANDOM_POPULATION Function (PV-WAVE Advantage)
Creates a population data structure from randomly generated individuals.
Usage
result = GA_RANDOM_POPULATION (n, chromosome)
Input Parameters
n—Scalar long value indicating the number of individuals to be randomly generated for the population.
chromosome—A
chromosome data structure created by the
GA_CHROMOSOME Function (PV-WAVE Advantage) describing the chromosome encoding for individuals.
Returned Value
result—GA_RANDOM_POPULATION returns a
population data structure, which is required input to
GENETIC_ALGORITHM Function (PV-WAVE Advantage).
Input Keywords
Print—If present and nonzero, this option turns on printing of intermediate results. By default, intermediate results are not printed.
Gray_encoding—If present and nonzero, specifies alleles are encoded using Gray encoding for integer and real phenotypes. If undefined or zero, specifies alleles are encoded using Base-2 encoding for integer and real phenotypes. Default: Base-2 encoding.
Pmx_crossover—This keyword applies partially matched crossover to the nominal portion of the chromosome. Although GA_RANDOM_POPULATION does not perform crossover in the population, this option signals that the nominal phenotypes are sequential with values consisting of an arrangement of the integers 0, 1, ..., N_nominal – 1, where N_nominal is the number of nominal phenotypes. Default: Standard crossover. Each nominal phenotype can independently have values from 0 to N_nominal – 1.
Fitness_fcn—User-supplied function to calculate fitness for individual. If this is supplied, fitness values are calculated for each individual and included within the expanded population data structure. Otherwise they are set to zero.
Binary_prob—The random selection model for randomly generating values for the binary phenotypes. By default binary phenotype values are selected with equal probability, i.e. p(0) = p (1) = 0.5. However, Binary_prob can be used to specify any Bernoulli distribution as the random selection model for individual binary phenotypes. Binary_prob is a one-dimensional array of length N_binary. Binary_prob(i) is equal to the probability that the ith binary phenotype equals zero. Hence the probability it equals one is 1 – Binary_prob(i).
Nominal_prob—The random selection model for randomly generating values for the nominal phenotypes are described by an array of length N_nominal, where:
By default all integer values between zero and
N_categories(
i) – 1 are selected with equal probability. However,
Nominal_prob can be used to specify any multinomial distribution as the random selection model for individual nominal phenotypes.
Nominal_prob is a jagged two dimensional array. The values in the
ith row of this array contain the probability of selecting 0, 1, ...,
N_categories(
i) – 1 for the
ith nominal attribute. These must be valid probabilities scaled between 0 and 1, and they must sum to 1.0. The number of values in the
ith row is equal to
N_categories(
i). See the
GA_CHROMOSOME Function (PV-WAVE Advantage) for a description of
N_categories.
Int_s_model—The random selection model for randomly generating values for the integer phenotypes. Int_s_model(i) declares the random selection model for the i th integer phenotype. If Int_s_model(i) = 0, all integer values between the upper and lower limits specified in chromosome, I_bounds(2i) and I_bounds(2i+1), are selected with equal probability. This is the default selection method for integer phenotypes. If Int_s_model(i) = 0, the contents of I_parms(2i) and I_parms(2i+1) are replaced with the lower limit of the interval and its width, respectively. If Int_s_model(i) = 1, the Poisson random selection model is used. The Poisson distribution models a population of non-negative integers. If this model is selected, then all values for the i th integer phenotype must be non-negative.
I_parms—An input/output array of size 2*chromosome.n_integer. If Int_s_model(i) is set to 0, then I_parms(2i) and I_parms(2i+1) are used to return the i th lower limit and width of the interval as described in the discussion of Int_s_model. If Int_s_model(i) is set to 1, then I_parms(2i) must contain the mean for the Poisson distribution for the i th integer phenotype and the value of I_parms(2i+1) is ignored.
Real_s_model—The random selection model for randomly generating values for the real phenotypes. Real_s_model(i) can be used to specify the random selection model for the ith real phenotype. If Real_s_model(i)= 0, all integer values between the upper and lower limits specified in chromosome, i_bounds(2i) and i_bounds(2i+1), are selected with equal probability. This is the default selection method for real phenotypes. If Real_s_model(i)= 0, the contents of R_parms(2i) and R_parms(2i+1) are replaced with the lower limit of the interval and its width, respectively. If Real_s_model(i)=1, then the Gaussian distribution is used. In this case, the value of R_parms(2i) should be set to the mean of this distribution and R_parms(2i+1) should equal its variance.
R_parms—An input/output array of size 2*chromosome.n_real. If Real_s_model(i) is set to 0, then R_parms(2i) and R_parms(2i+1) are used to return the i th lower limit and width of the interval as described in the discussion of Real_s_model. If Real_s_model(i) is set to 1, then R_parms(2i) must contain the mean for the Gaussian distribution for the i th integer phenotype and R_parms(2i+1) should equal its variance.
Double—If present and nonzero, then double precision is used.
Discussion
The
GENETIC_ALGORITHM Function (PV-WAVE Advantage) operates on a population of individuals. GA_RANDOM_POPULATION creates an initial population of
n randomly selected individuals. GA_RANDOM_POPULATION takes the chromosome structure described by the
chromosome argument and randomly generates values for each phenotype. These are then encoded into
individual data structures and placed into the population.
Binary phenotypes are randomly generated Bernoulli random variables with p(0) = p(1)=0.5.
Values for nominal phenotypes are generated with equal probability. That is the probability of sampling each of the N_categories(i) values for the ith nominal phenotype is 1/(chromosome.N_categories(i)).
By default, random values for the integer phenotypes are generated using the discrete uniform distribution. All values between I_bounds(2i) and I_bounds(2i+1) are sampled with equal probability. This default can be changed using the Int_s_model keyword.
Likewise, random values for real phenotypes are generated using the continuous uniform random distribution. All values between R_bounds(2i) and R_bounds(2i+1) are sampled with equal probability. This default can be changed using the Real_s_model keyword.
Example
This example creates a population of 40 individuals each with with 1 binary, 2 nominals, 3 integers and 2 real phenotypes. The
Print keyword is used to print a description of the population. A simple fitness function calculation is used to illustrate how fitness values can be used to initialize a population with the
Fitness_fcn keyword. If fitness is not initialized, the fitness array in the data structure is set to null. It can be initialized using a keyword with
GENETIC_ALGORITHM Function (PV-WAVE Advantage).
PRO ga_random_population_ex1
; Number of phenotypes by category.
n_binary = 1
n_nominal = 2
n_integer = 3
n_real = 2
n = 40 ; Population size
binaryPhenotype = [1] ; binary phenotype
; Number of categories for nomial phenotypes.
n_categories = [2, 3]
; Nominal phenotype values
nominalPhenotype = [1, 2]
; Number of intervals and boundaries for integer
; phenotypes.
i_intervals = [16, 16, 16]
i_bounds = [0, 1000, -10, 10, -20, 0]
; Integer phenotype values.
integerPhenotype = [200, -5, -5]
; Number of intervals and boundaries for real
; phenotypes.
r_intervals = [512, 1024]
r_bounds = [0.0, 20.0, -20.0, 20.0]
; Real phenotype values.
realPhenotype = [19.9, 19.9]
; Fitness array for individuals.
fitness = FLTARR(40)
; The individuals in the population.
individuals = LISTARR(n)
chromosome = GA_CHROMOSOME( $
N_binary=n_binary, $
N_nominal=n_nominal, $
N_categories=n_categories, $
N_integer=n_integer, $
I_intervals=i_intervals, $
I_bounds=i_bounds, $
N_real=n_real, $
R_intervals=r_intervals, $
R_bounds=r_bounds)
; Create reproducible results.
RANDOMOPT,set=12345
population = GA_RANDOM_POPULATION(40, chromosome,$
Fitness_fcn="at_fitness",/Print)
END
FUNCTION at_fitness, individual
; Calculate fitness for this individual.
fitness = 100.0 + 10*individual.binary_Phenotype(0)
fitness = fitness + 2*individual.nominal_Phenotype(1) - $
4*individual.nominal_Phenotype(0)
fitness = fitness + 0.0001*individual.integer_Phenotype(0) + $
ABS(individual.integer_Phenotype(1)+ $
individual.integer_Phenotype(2))*0.1
fitness = fitness + 0.1*individual.real_Phenotype(0) + $
ABS(individual.real_Phenotype(1))*0.2
RETURN, fitness
END
Output
The Print option produced the following description of the population. A summary of the population chromosome structure and fitness are printed followed by detailed information for the first 5 individuals in the population.
This example also illustrates the bit ordering within chromosomes. Nominal phenotypes are placed in the first bits followed by binary and encoded integer and real phenotypes. Note that this output is identical to the example for GA_POPULATION because the fitness function is identical and the random phenotype generation uses the same random seed.
Population Size: 40
Average Fitness: 109.400070
Std. Dev. Fitness: 5.923696
Maximum Fitness: 120.044495
Minimum Fitness: 98.022018
Chromosome:
*******************************
**** CHROMOSOME STRUCTURE *****
Chromosome length: 34 Bits
*****BIT ASSIGNMENTS***********
Binary: 2 - 2 n_binary = 1
Nominal: 1 - 2 n_nominal= 2
Integer: 3 - 14 n_integer= 3
Real: 15 - 33 n_real = 2
*******************************
NOMINAL CATEGORIES*************
Variable 0: 2 categories
Variable 1: 3 categories
*******************************
INTEGER BOUNDS*****************
Variable 0: [ 0, 1000]
Variable 1: [ -10, 10]
Variable 2: [ -20, 0]
*******************************
INTEGER BITS*******************
Variable 0: 4 bits
Variable 1: 4 bits
Variable 2: 4 bits
*******************************
INTEGER DISCRETE INTERVALS*****
Variable 0: 16 intervals
Variable 1: 16 intervals
Variable 2: 16 intervals
*******************************
REAL BOUNDS********************
Variable 0: [0,20]
Variable 1: [-20,20]
*******************************
REAL BITS**********************
Variable 0: 9 bits
Variable 1: 10 bits
*******************************
REAL DISCRETE INTERVALS********
Variable 0: 512 intervals
Variable 1: 1024 intervals
*******************************
First 5 Individuals
****** INDIVIDUAL 0 ********************************************
Number of Parents: 2
Encoding: BASE-2
Fitness: 105.114510
PHENOTYPES
*************BINARY************
Variable 0: 0
************NOMINAL************
Variable 0: 1
Variable 1: 2
************INTEGER************
Variable 0: 35
Variable 1: -10
Variable 2: -19
**************REAL*************
Variable 0: 15.3157
Variable 1: 3.39719
**********CHROMOSOME*******************************************
1 2 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 0 1 0 0 0 1 0 0 1 0 1 0 1 1 0
***************************************************************
****** INDIVIDUAL 1 ********************************************
Number of Parents: 2
Encoding: BASE-2
Fitness: 111.696175
PHENOTYPES
*************BINARY************
Variable 0: 1
************NOMINAL************
Variable 0: 1
Variable 1: 0
************INTEGER************
Variable 0: 195
Variable 1: -5
Variable 2: -4
**************REAL*************
Variable 0: 19.6777
Variable 1: -14.0445
**********CHROMOSOME*******************************************
1 0 1 0 0 1 1 0 1 0 0 1 1 0 0 1 1 1 1 1 0 1 1 1 0 0 1 0 0 1 1 0 0 0
***************************************************************
****** INDIVIDUAL 2 ********************************************
Number of Parents: 2
Encoding: BASE-2
Fitness: 104.741798
PHENOTYPES
*************BINARY************
Variable 0: 0
************NOMINAL************
Variable 0: 0
Variable 1: 0
************INTEGER************
Variable 0: 167
Variable 1: 8
Variable 2: -16
**************REAL*************
Variable 0: 18.3331
Variable 1: -10.4589
**********CHROMOSOME*******************************************
0 0 0 0 0 1 0 1 1 1 0 0 0 1 1 1 1 1 0 1 0 1 0 1 0 0 1 1 1 1 0 1 0 0
***************************************************************
****** INDIVIDUAL 3 ********************************************
Number of Parents: 2
Encoding: BASE-2
Fitness: 110.805908
PHENOTYPES
*************BINARY************
Variable 0: 1
************NOMINAL************
Variable 0: 1
Variable 1: 0
************INTEGER************
Variable 0: 630
Variable 1: 0
Variable 2: -16
**************REAL*************
Variable 0: 18.213
Variable 1: -6.608
**********CHROMOSOME*******************************************
1 0 1 1 0 1 0 1 0 0 0 0 0 1 1 1 1 1 0 1 0 0 1 0 0 1 0 1 0 1 0 1 1 0
***************************************************************
****** INDIVIDUAL 4 ********************************************
Number of Parents: 2
Encoding: BASE-2
Fitness: 114.571022
PHENOTYPES
*************BINARY************
Variable 0: 1
************NOMINAL************
Variable 0: 1
Variable 1: 2
************INTEGER************
Variable 0: 51
Variable 1: 9
Variable 2: -2
**************REAL*************
Variable 0: 7.13049
Variable 1: -15.7644
**********CHROMOSOME*******************************************
1 2 1 0 0 0 0 1 1 1 1 1 1 1 0 0 1 0 1 1 0 1 1 0 0 0 0 1 1 0 1 1 0 0
***************************************************************
Version 2017.0
Copyright © 2017, Rogue Wave Software, Inc. All Rights Reserved.