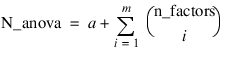
x = MACHINE(/Float)
y(i) = x.NaN
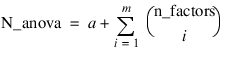
J | anova_tablei,j = anova_table(i,j) |
0 | Source Identifier (values described below) |
1 | Degrees of freedom |
2 | Sum of squares |
3 | Mean squares |
4 | F-statistic |
5 | p-value for this F-statistic |
Source Identifier | ANOVA Source |
–1 | Main Effects1 |
–2 | Two-Way Interactions2 |
–3 | Three-Way Interactions2 |
. | . |
. | . |
. | . |
–n_factors | (n_factors)-way Interactions2 |
–n_factors – 1 | Effects Error Term |
–n_factors – 2 | Residual3 |
–n_factors – 3 | Corrected Total |
Notes: 1. The number of main effects is equal to n_factors + 1 if n_locations > 1, and n_factors if n_locations = 1. The first row of values, anova_table(0,0) through anova_table(0,5) contain the location effect if n_locations > 1. If n_locations = 1, then these values are the effects for factor 1. 2. The number of interaction effects for the nth-way interactions is equal to: 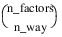 The order of these terms is in ascending order by treatment subscript. The interactions for factor 1 appear first, followed by factor 2, factor 3, and so on. 3. The residual term is only provided when treatments are replicated, i.e., n_levels(n_factors) > 1. | |
note | By default, model_order = n_factors when treatments are replicated, or n_locations > 1. However, if treatments are not replicated and n_locations = 1, model_order = n_factors – 1. |
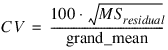
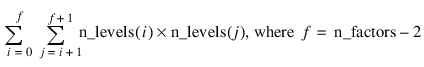
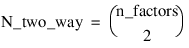
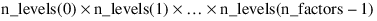
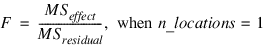
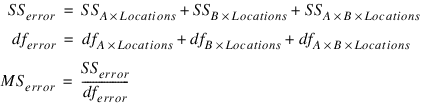
n_obs = 12
n_locations = 1
n_factors = 3
n_levels = [3, 2, 2, 1]
models = TRANSPOSE( $
[ [1, 1, 1, 1], $
[1, 1, 1, 2], $
[1, 1, 2, 1], $
[1, 1, 2, 2], $
[1, 2, 1, 1], $
[1, 2, 1, 2], $
[1, 2, 2, 1], $
[1, 2, 2, 2], $
[1, 3, 1, 1], $
[1, 3, 1, 2], $
[1, 3, 2, 1], $
[1, 3, 2, 2] ])
; Response data
y = [4.42725419998168950, $
2.12795543670654300, $
2.55254390835762020, $
1.21479606628417970, $
2.47588264942169190, $
5.01306104660034180, $
4.73502767086029050, $
4.58392113447189330, $
5.01421167794615030, $
4.11972457170486450, $
6.51671624183654790, $
4.73365202546119690]
a = MACHINE(/Float)
NaN = a.NAN
y(8) = NaN
aov = CRD_FACTORIAL(n_obs, n_locations, n_factors, $
n_levels, models, y, $
N_missing=n_missing, $
Cv=cv, Grand_mean=grand_mean, $
Factor_means=factor_means, $
Factor_std_errors=factor_std_errors, $
Two_way_means=two_way_means, $
Two_way_std_errors=two_way_std_errors, $
Treatment_means=treatment_means, $
Treatment_std_errors=treatment_std_errors, $
Anova_row_labels=anova_row_labels)
; Print Analysis of Variance Table
PRINT, " *** ANALYSIS OF VARIANCE TABLE ***"
PRINT, 'ID', 'DF', 'SSQ', 'MS', 'F-Test', 'p-Value', $
Format='(A12, A6, A8, A8, A8, A9)'
FOR i=0L, (SIZE(aov))(1)-1 DO BEGIN & $
PRINT, anova_row_labels(i), aov(i,0), aov(i,1), $
aov(i,2), aov(i,3), aov(i,4), aov(i,5), Format= $
'(A7, 2X, I3, 3X, F3.0, 2X, F6.2, 2X, F6.2, 2X, ' + $
'F6.2, 4X, F5.3)' & $
ENDFOR
PRINT, ''
PRINT, n_missing, $
Format='("Number of Missing Values Estimated:", I3)'PRINT, grand_mean, $
Format='("Grand Mean :", F7.3)'PRINT, cv, $
Format='("Coefficient of Variation :", F7.3)'PRINT, ''
l = 0 & m = 0
PRINT, "Factor Means"
FOR i=0L, n_factors-1 DO BEGIN & $
PRINT, (i+1), factor_means(m:(m+n_levels(i)-1)), $
Format='(2X, "Factor", I2, ": ", ' + $
STRTRIM(n_levels(i),2) + 'F11.6)' & $
k = FIX(factor_std_errors(l,1)) & $
PRINT, factor_std_errors(i), k, $
Format='(15X, "std. err.(df):", F16.6, "(", I1, ")")' & $l = l + 1 & $
m = m + n_levels(i) & $
ENDFOR
PRINT, ''
l = 0 & m = 0
PRINT, "Two-Way Means"
FOR i=0L, n_factors-1 DO BEGIN & $
FOR j=(i+1), n_factors-1 DO BEGIN & $
PRINT, (i+1), (j+1), Format= $
'(2X, "Factor", I2, " by Factor", I2, ":")' & $
FOR i2=0L, (n_levels(i)-1) DO BEGIN & $
PRINT, two_way_means(m:(m+n_levels(j)-1)) & $
m = m + n_levels(j) & $
ENDFOR & $
k = FIX(two_way_std_errors(l,1)) & $
PRINT, two_way_std_errors(l), k, $
Format='(2X, "std. err.(df): = ", F7.5, ' + $
'"(", I1, ")")' & $l = l + 1 & $
PRINT, '' & $
ENDFOR & $
ENDFOR
PRINT, "Treatment Means"
m = 0
FOR i=0L, n_levels(0)-1 DO $
FOR j=0L, n_levels(1)-1 DO $
FOR k=0L, n_levels(2)-1 DO BEGIN & $
PRINT, (i+1), (j+1), (k+1), treatment_means(m), Format= $
"(2X, 'Treatment[', I1, '][', I1, '][', I1, ']', " + $
"' Mean:', F7.4)" & $
m = m + 1 & $
ENDFOR
PRINT, ''
k = FIX(Treatment_std_errors(1)) & $
PRINT, Treatment_std_errors(0), k, $
Format='("Treatment Std. Err.(df): = ", F7.5, "(", I1, ")")'*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-test P-Value
[1] -1 2. 13.06 6.53 7.84 0.245
[2] -1 1. 0.11 0.11 0.13 0.781
[3] -1 1. 1.30 1.30 1.56 0.429
[1]x[2] -2 2. 3.77 1.88 2.26 0.425
[1]x[3] -2 2. 5.25 2.63 3.15 0.370
[2]x[3] -2 1. 0.56 0.56 0.67 0.563
Error -4 1. 1.67 1.67 NaN NaN
Total -5 10. 25.72 NaN NaN NaN
Number of Missing Values Estimated: 1
Grand Mean : 3.962
Coefficient of Variation : 32.574
Factor Means
Factor 1: 2.580637 4.201973 5.101930
std. err.(df): 0.912459(1)
Factor 2: 3.866917 4.056109
std. err.(df): 0.745020(1)
Factor 3: 4.290842 3.632185
std. err.(df): 0.745020(1)
Two-Way Means
Factor 1 by Factor 2:
3.27760 1.88367
3.74447 4.65947
4.57868 5.62518
std. err.(df): = 1.29041(1)
Factor 1 by Factor 3:
3.48990 1.67138
3.60546 4.79849
5.77717 4.42669
std. err.(df): = 1.29041(1)
Factor 2 by Factor 3:
3.98025 3.75358
4.60143 3.51079
std. err.(df): = 1.05362(1)
Treatment Means
Treatment[1][1][1] Mean: 4.4273
Treatment[1][1][2] Mean: 2.1280
Treatment[1][2][1] Mean: 2.5525
Treatment[1][2][2] Mean: 1.2148
Treatment[2][1][1] Mean: 2.4759
Treatment[2][1][2] Mean: 5.0131
Treatment[2][2][1] Mean: 4.7350
Treatment[2][2][2] Mean: 4.5839
Treatment[3][1][1] Mean: 5.0376
Treatment[3][1][2] Mean: 4.1197
Treatment[3][2][1] Mean: 6.5167
Treatment[3][2][2] Mean: 4.7337
Treatment Std. Err.(df): = 1.82492(1)