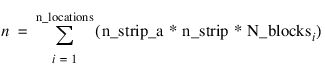
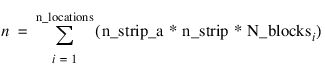
x = MACHINE(/Float)
y(i) = x.NaN
J | anova_tablei,j = anova_table(i,j) |
0 | Source Identifier (values described below) |
1 | Degrees of freedom |
2 | Sum of squares |
3 | Mean squares |
4 | F-statistic |
5 | p-value for this F-statistic |
Source Identifier | ANOVA Source |
-1 | LOCATIONS* |
-2 | BLOCK WITHIN LOCATION |
-3 | STRIP-PLOT A |
-4 | LOCATION × STRIP-PLOT A* |
-5 | STRIP-PLOT A ERROR |
-6 | SPLIT-PLOT |
-7 | SPLIT-PLOT × STRIP-PLOT A |
-8 | LOCATION × SPLIT-PLOT* |
-9 | SPLIT-PLOT ERROR |
-10 | LOCATION × SPLIT-PLOT × STRIP-PLOT A* |
-11 | STRIP-PLOT B |
-12 | LOCATION × STRIP-PLOT B* |
-13 | STRIP_PLOT B ERROR |
-14 | STRIP-PLOT A × STRIP-PLOT B |
-15 | LOCATION × STRIP-PLOT A × STRIP-PLOT B |
-16 | STRIP-PLOT A × STRIP-PLOT B ERROR |
-17 | SPLIT-PLOT × STRIP-PLOT B |
-18 | STRIP-PLOT A × STRIP-PLOT B × SPLIT-PLOT |
-19 | LOCATION × SPLIT-PLOT × STRIP-PLOT B* |
-20 | LOCATION × STRIP-PLOT A × STRIP-PLOT B × SPLIT-PLOT* |
-21 | STRIP-PLOT A × STRIP-PLOT B × SPLIT-PLOT ERROR |
-22 | CORRECTED TOTAL |
* If n_locations = 1 sources involving location are set to missing (NaN). | |
Element | Standard Error for Comparisons Between Two | Degrees of Freedom |
Std_errors(0) | Strip-Plot A Means | Std_errors(10) |
Std_errors(1) | Strip-Plot B Means | Std_errors(11) |
Std_errors(2) | Split-Plot Means | Std_errors(12) |
Std_errors(3) | Strip-Plot A Means at the same level of split-plots | Std_errors(13) |
Std_errors(4) | Strip-Plot A Means at the same level of strip-plot B | Std_errors(14) |
Std_errors(5) | Strip-Plot B Means at the same level of split-plots | Std_errors(15) |
Std_errors(6) | Strip-Plot B Means at the same level of strip-plot A | Std_errors(16) |
Std_errors(7) | Split-Plot Means at the same level of split-plot A | Std_errors(17) |
Std_errors(8) | Split-Plot Means at the same level of strip-plot B | Std_errors(18) |
Std_errors(9) | Treatment Means (same strip-plot A, strip-plot B and split-plot) | Std_errors(19) |
Factor A Strip-Plots | |||||
A2 | A1 | A4 | A3 | ||
Factor B Strip Plots | B3 | A2B3 | A1B3 | A4B3 | A3B3 |
B1 | A2B1 | A1B1 | A4B1 | A3B1 | |
B2 | A2B2 | A1B2 | A4B2 | A3B2 | |
Whole-Plot Factor | |||
A2 | A1 | A4 | A3 |
A2B1 | A1B3 | A4B1 | A3B3 |
A2B3 | A1B1 | A4B3 | A3B1 |
A2B2 | A1B2 | A4B2 | A3B2 |
Factor A Strip Plots | |||||
A2 | A1 | A4 | A3 | ||
Factor B Strip Plots | B3 | A2B3C2 A2B3C1 | A1B3C1 A1B3C2 | A4B3C2 A4B3C1 | A3B3C2 A3B3C1 |
B1 | A2B1C1 A2B1C2 | A1B1C1 A1B1C2 | A4B1C2 A4B1C1 | A3B1C2 A3B1C1 | |
B2 | A2B2C2 A2B2C1 | A1B2C1 A1B2C2 | A4B2C1 A4B2C2 | A3B2C2 A3B2C1 | |
Whole Plot Factor A | |||
A2 | A1 | A4 | A3 |
A2B3C2 A2B3C1 | A1B2C1 A1B2C2 | A4B1C2 A4B1C1 | A3B3C2 A3B3C1 |
A2B1C1 A2B1C2 | A1B1C1 A1B1C2 | A4B3C2 A4B3C1 | A3B2C2 A3B2C1 |
A2B2C2 A2B2C1 | A1B3C1 A1B3C2 | A4B2C1 A4B2C2 | A3B1C2 A3B1C1 |
Fertilizer Strip Plots | |||||
F2 | F1 | F4 | F3 | ||
Application Rate Strip Plot | R3 | F2R3S1 F2R3S2 F2R3S3 | F1R3S3 F1R3S2 F1R3S1 | F4R3S3 F4R3S2 F4R3S1 | F3R3S2 F3R3S1 F3R3S3 |
R2 | F2R1S3 F2R1S1 F2R1S2 | F1R1S2 F1R1S3 F1R1S1 | F4R1S3 F4R1S1 F4R1S2 | F3R1S1 F3R1S2 F3R1S3 | |
R1 | F2R2S1 F2R2S2 F2R2S3 | F1R2S1 F1R2S3 F1R2S2 | F4R2S2 F4R2S3 F4R2S1 | F3R2S3 F3R2S1 F3R2S2 | |
; Total number of observations
n = 24
; Number of locations
n_locations = 1
; Number of factor A strip-plots within a location
n_strip_a = 2
; Number of factor B strip-plots within a location
n_strip_b = 2
; Number of split-plots within each Factor A strip-plot
n_split = 2
block = [1, 1, 1, 1, 1, 1, 1, 1, $
2, 2, 2, 2, 2, 2, 2, 2, $
3, 3, 3, 3, 3, 3, 3, 3]
strip_a = [1, 1, 1, 1, 2, 2, 2, 2, $
1, 1, 1, 1, 2, 2, 2, 2, $
1, 1, 1, 1, 2, 2, 2, 2]
strip_b = [1, 1, 2, 2, 1, 1, 2, 2, $
1, 1, 2, 2, 1, 1, 2, 2, $
1, 1, 2, 2, 1, 1, 2, 2]
split = [1, 2, 1, 2, 1, 2, 1, 2, $
1, 2, 1, 2, 1, 2, 1, 2, $
1, 2, 1, 2, 1, 2, 1, 2]
y = [30.0, 40.0, 38.9, 38.2, $
41.8, 52.2, 54.8, 58.2, $
20.5, 26.9, 21.4, 25.1, $
26.4, 36.7, 28.9, 35.9, $
21.0, 25.4, 24.0, 23.3, $
34.4, 41.0, 33.0, 34.9]
aov = STRIP_SPLIT_PLOT(n, n_locations, n_strip_a, $
n_strip_b, n_split, block, $
strip_a, strip_b, split, y, $
Cv=cv, Grand_mean=grand_mean, $
Treatment_means=treatment_means, $
Strip_plot_a_means= $
strip_plot_a_means, $
Strip_plot_b_means=$
strip_plot_b_means, $
Split_plot_means=split_plot_means, $
Strip_plot_a_split_plot_means= $
strip_plot_a_split_plot_means, $
Strip_plot_b_split_plot_means= $
strip_plot_b_split_plot_means, $
Strip_plot_ab_means= $
strip_plot_ab_means, $
Std_errors=std_errors, $
N_blocks=n_blocks, $
Location_anova_table= $
location_anova_table)
labels = ['Location ', $
'Blocks ', $
'Strip-Plot A ', $
'Location x A ', $
'Strip-Plot A Error ', $
'Split-Plot ', $
'Split-Plot x A ', $
'Location x ', $
' Split-Plot ', $
'Split-Plot Error ', $
'Location x ', $
' Split-Plot x A ', $
'Strip-Plot B ', $
'Location x B ', $
'Strip-Plot B Error ', $
'A x B ', $
'Location x A x B ', $
'A x B Error ', $
'Split-Plot x B ', $
'Split-Plot x A x B ', $
'Location x ', $
' Split-Plot x B ', $
'Location x ', $
' Split-Plot x A x B', $
'Split-Plot x A x ', $
' B Error ', $
'Corrected Total ']
; Print Analysis of Variance Table
PRINT, " *** ANALYSIS OF VARIANCE TABLE ***"
PRINT, 'ID', 'DF', 'SSQ', 'MS', 'F-Test', 'p-Value', $
Format='(A24, A5, A9, A8, A7, A8)'
idx = 0
FOR i=0L, (SIZE(aov))(1)-1 DO BEGIN & $
PRINT, labels(idx), aov(i,0), aov(i,1), aov(i,2), $
aov(i,3), aov(i,4), aov(i,5), Format= $
'(A20, 1X, I3, 2X, F3.0, 2X, F7.2, 2X, F6.2, 2X, ' + $
'F5.2, 2X, F6.3)' & $
idx = idx + 1 & $
IF idx LT N_ELEMENTS(labels)-1 THEN $
WHILE STRPOS(labels(idx), ' ', 0) EQ 0 DO BEGIN & $
PRINT, labels(idx) & idx = idx + 1 & $
ENDWHILE & $
ENDFOR
PRINT, ''
PRINT, grand_mean, Format="('Grand mean: ', F9.6, '\012')"PRINT, "Coefficient of Variation"
labels = ['Strip-Plot A:', 'Strip-Plot B:','Strip-Plot: ']
FOR i=0L, 2 DO PRINT, labels(i), cv(i), Format='(A15, F12.4)'
PRINT, ''
PRINT, "****************************************************"
; Print the Treatment Means
PRINT, "Treatment Means"
idx = 0
FOR i=0L, n_strip_a-1, idx+1 DO $
FOR j=0L, n_strip_b-1 DO $
FOR k=0L, n_split-1 DO $
PRINT, (i+1), (j+1), (k+1), $
treatment_means(i,j,k), Format= "('" + $" Treatment[', I1, '][', I1, '][', I1, '] Mean:', F9.4)"
PRINT, ''
PRINT, std_errors(9), $
Format='("Standard Error for Comparing Two ' + $'Treatment Means:", F9.6)'
PRINT, std_errors(19), Format="('(df=', F8.6, ')', '\012')"; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(REFORM(treatment_means, $
(n_strip_a*n_strip_b*n_split)), $
std_errors(19), std_errors(9)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, "****************************************************"
; Print the Strip-plot A Means.
;
PM, strip_plot_a_means, Title="Strip-plot A means"
PRINT, ''
PRINT, std_errors(0), $
Format='("Standard Error for Comparing Two ' + $'Strip-Plot A Means:", F9.6)'
PRINT, std_errors(10), Format="('(df=', F8.6, ')', '\012')"; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(strip_plot_a_means, $
std_errors(10), std_errors(0)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, "****************************************************"
; Print the Strip-plot B Means.
;
PM, strip_plot_b_means, Title="Strip-plot B means"
PRINT, ''
PRINT, std_errors(1), $
Format='("Standard Error for Comparing Two ' + $'Strip-Plot B Means:", F9.6)'
PRINT, std_errors(11), Format="('(df=', F8.6, ')', '\012')"; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(strip_plot_b_means, $
std_errors(11), std_errors(1)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, "****************************************************"
; Print the Split-plot Means.
PM, split_plot_means, Title="Split-plot Means"
PRINT, ''
PRINT, std_errors(2), $
Format='("Standard Error for Comparing Two ' + $'Split-Plot Means:", F9.6)'
PRINT, std_errors(12), Format="('(df=', F8.6, ')', '\012')"; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(split_plot_means, $
std_errors(12), std_errors(2)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, "****************************************************"
; Print the Strip-plot A by Split-plot Means.
PM, strip_plot_a_split_plot_means, $
Title="Strip-plot A by Split-plot Means"
PRINT, ''
PRINT, std_errors(3), $
Format='("Standard Error for Comparing Two Means:", F9.6)'PRINT, std_errors(13), Format="('(df=', F8.6, ')', '\012')"; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP( $
REFORM(strip_plot_a_split_plot_means, $
n_strip_a*n_split), $
std_errors(13), std_errors(3)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, "****************************************************"
; Print the Strip-plot A by Split-plot B Means.
;
PM, strip_plot_ab_means, $
Title="Strip-plot A by Strip-plot B Means"
PRINT, ''
PRINT, std_errors(4), $
Format='("Standard Error for Comparing Two Means:", F9.6)'PRINT, std_errors(14), Format="('(df=', F8.6, ')', '\012')"; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP(REFORM(strip_plot_ab_means, $
n_strip_a*n_strip_b), $
std_errors(14), std_errors(4)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
PRINT, "****************************************************"
; Print the Strip-plot B by Split-plot Means.
PM, strip_plot_b_split_plot_means, $
Title="Strip-plot B by Split-plot Means"
PRINT, ''
PRINT, std_errors(5), $
Format='("Standard Error for Comparing Two Means:", F9.6)'PRINT, std_errors(15), Format="('(df=', F8.6, ')', '\012')"PRINT, ''
; Perform multiple comparison using the LSD procedure
equal_means = MULTICOMP($
REFORM(strip_plot_b_split_plot_means, $
n_strip_b*n_split), $
std_errors(15), std_errors(5)/SQRT(2.0), $
/LSD, Alpha=0.05)
PM, equal_means, $
Title="LSD Comparison: Size of Groups of Means"
PRINT, ''
*** ANALYSIS OF VARIANCE TABLE ***
ID DF SSQ MS F-Test p-Value
Location -1 NaN NaN NaN NaN NaN
Blocks -2 2. 1310.28 655.14 14.53 0.061
Strip-Plot A -3 1. 858.01 858.01 40.37 0.024
Location x A -4 NaN NaN NaN NaN NaN
Strip-Plot A Error -5 2. 42.51 21.26 1.48 0.385
Split-Plot -6 1. 163.80 163.80 41.22 0.003
Split-Plot x A -7 1. 11.34 11.34 2.85 0.166
Location x -8 NaN NaN NaN NaN NaN
Split-Plot
Split-Plot Error -9 4. 15.90 3.97 1.56 0.338
Location x -10 NaN NaN NaN NaN NaN
Split-Plot x A
Strip-Plot B -11 1. 17.17 17.17 0.47 0.565
Location x B -12 NaN NaN NaN NaN NaN
Strip-Plot B Error -13 2. 73.51 36.75 2.85 0.260
A x B -14 1. 1.55 1.55 0.12 0.762
Location x A x B -15 NaN NaN NaN NaN NaN
A x B Error -16 2. 25.82 12.91 5.08 0.080
Split-Plot x B -17 1. 46.76 46.76 18.39 0.013
Split-Plot x A x B -18 1. 0.51 0.51 0.20 0.677
Location x -19 NaN NaN NaN NaN NaN
Split-Plot x B
Location x -20 NaN NaN NaN NaN NaN
Split-Plot x A x B
Split-Plot x A x -21 4. 10.17 2.54 NaN NaN
B Error
Corrected Total -22 23. 2577.33 NaN NaN NaN
Grand mean: 33.870834
Coefficient of Variation
Strip-Plot A: 13.6116
Strip-Plot B: 17.8986
Strip-Plot: 5.8854
****************************************************
Treatment Means
Treatment[1][1][1] Mean: 23.8333
Treatment[1][1][2] Mean: 30.7667
Treatment[1][2][1] Mean: 28.1000
Treatment[1][2][2] Mean: 28.8667
Treatment[2][1][1] Mean: 34.2000
Treatment[2][1][2] Mean: 43.3000
Treatment[2][2][1] Mean: 38.9000
Treatment[2][2][2] Mean: 43.0000
Standard Error for Comparing Two Treatment Means: 1.302029
(df=4.000000)
LSD Comparison: Size of Groups of Means
0
3
0
2
0
0
2
****************************************************
Strip-plot A means
27.8917
39.8500
Standard Error for Comparing Two Strip-Plot A Means: 1.882171
(df=2.000000)
LSD Comparison: Size of Groups of Means
0
****************************************************
Strip-plot B means
33.0250
34.7167
Standard Error for Comparing Two Strip-Plot B Means: 2.474972
(df=2.000000)
LSD Comparison: Size of Groups of Means
2
****************************************************
Split-plot Means
31.2583
36.4833
Standard Error for Comparing Two Split-Plot Means: 0.813813
(df=4.000000)
LSD Comparison: Size of Groups of Means
0
****************************************************
Strip-plot A by Split-plot Means
25.9667 29.8167
36.5500 43.1500
Standard Error for Comparing Two Means: 1.150906
(df=4.000000)
LSD Comparison: Size of Groups of Means
0
0
0
****************************************************
Strip-plot A by Strip-plot B Means
27.3000 28.4833
38.7500 40.9500
Standard Error for Comparing Two Means: 2.074280
(df=2.000000)
LSD Comparison: Size of Groups of Means
2
0
2
****************************************************
Strip-plot B by Split-plot Means
29.0167 37.0333
33.5000 35.9333
Standard Error for Comparing Two Means: 0.920673
(df=4.000000)
LSD Comparison: Size of Groups of Means
0
2
2